Machine Learning on Apple stock daily return2
Tags: IBM, Machine Learning
Categories: Projects
Updated:
import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
%matplotlib inline
import seaborn as sns
1. Data
aapl = pd.read_csv('./data/aapl.csv')
X = aapl.drop(['Date','daily_ret'], axis=1)
y = aapl['daily_ret']
aapl.columns
Index(['Date', 'Close', 'Volume', 'Lo', 'HO', 'CO', 'support_low',
'support_open', 'support_high', 'std20', 'std120', 'std_open20',
'std_high20', 'std_intra', 'ma20', 'ma120', 'daily_ret', 'd_mom',
'3d_mom', '5d_mom', '20d_mom', '252d_mom', 'AAPL_factor2',
'AAPL_factor1', 'change_wti', 'Lo_wti', 'HO_wti', 'CO_wti',
'change_nasdaq', 'Lo_nasdaq', 'HO_nasdaq', 'CO_nasdaq',
'Close_bond_10y', 'high6m_bond_10y', 'Close_bond_2y', 'high6m_bond_2y',
'change_itw', 'Lo_itw', 'HO_itw', 'CO_itw', 'Close_bond_1m',
'high6m_bond_1m', 'Close_bond_1y', 'high6m_bond_1y', 'Close_vix',
'change_vix', 'Lo_vix', 'HO_vix', 'CO_vix', 'high6m_vix', 'high1y_vix',
'Close_sp500', 'change_sp500', 'Lo_sp500', 'HO_sp500', 'CO_sp500',
'high6m_sp500', 'high1y_sp500', 'y', 'm', 'd', 'vix_cat',
'vix_change_cat', 'sp500_cat', 'sp500_change_cat'],
dtype='object')
1-1 Determining Normality
BoxCox Transformation
## from scipy.stats.mstats import normaltest ## D'Agostino K^2 Test
## from scipy.stats import boxcox ## Not doing this since it's time series data and not normally distributed
## ## Adding boxcox value
## aapl_cut = aapl.drop(['Date','daily_ret'], axis=1)
## nomres = normaltest(aapl_cut)
## aapl_cut = aapl_cut.iloc[:,np.where(nomres[0] > 0.05)[0].tolist()]
## for i in aapl_cut.columns:
## bc_res = boxcox(aapl_cut[i])
## aapl['box_'+i] = bc_res[0]
Using dummy variables
Making dummy variable based on categorized data
- vix, vix change
- sp500, sp500 change
opt = True
if opt:
one_hot_encode_cols = aapl.dtypes[aapl.dtypes == aapl.vix_cat.dtype] ## filtering by string categoricals
one_hot_encode_cols = one_hot_encode_cols.index.tolist() ## list of categorical fields
## Do the one hot encoding
df = pd.get_dummies(aapl, columns=one_hot_encode_cols, drop_first=True).reset_index(drop=True)
## ## Combination of dummy variables: Not useful since it takes a lot of computing power
## pd.options.mode.chained_assignment = None
## temp = pd.DataFrame()
## for i in [one_hot_encode_cols[0],one_hot_encode_cols[2]]:
## lists = df.columns[1:]
## for name in lists:
## if name in "daily_ret":
## continue
## if name[:-2] not in one_hot_encode_cols:
## for j in range(2,5):
## temp[name+'*'+i+'_'+str(j)] = df[name]*df[i+'_'+str(j)]
## df = pd.concat([df, temp], axis=1)
else:
df = aapl
def singular_test(data):
dc = data.corr()
non = (dc ==1).sum() !=1
return dc[non].T[non]
singular_test(df)
2. Sklearn learing model
from sklearn.preprocessing import StandardScaler, PolynomialFeatures
from sklearn.metrics import r2_score, confusion_matrix, classification_report, \
accuracy_score, precision_score, recall_score, f1_score
from sklearn.model_selection import TimeSeriesSplit, cross_validate, \
RandomizedSearchCV, GridSearchCV, train_test_split
from sklearn.pipeline import Pipeline
Testing learning model
def evaluate(model, X, y, cv,
scoring = ['r2', 'neg_mean_squared_error'],
validate = True):
if validate:
verbose = 0
else:
verbose = 2
scoring = ['r2', 'neg_mean_squared_error']
cv_results = cross_validate(model, X, y, cv=cv,
scoring = scoring, verbose=verbose)
return cv_results
def regress_test(data, regressor, params = None,
target ='daily_ret', window = 120, pred_window = 30):
## training with 6month(120days) and predict 3month(60days)
X = data.drop([target], axis=1)
y = data[target]
tscv = TimeSeriesSplit() ## n_splits=_num_batch
pf = PolynomialFeatures(degree=1)
alphas = np.geomspace(50, 800, 20)
scores=[]
for alpha in alphas:
ridge = Ridge(alpha=alpha, max_iter=100000)
estimator = Pipeline([
('scaler', StandardScaler()),
("polynomial_features", pf),
("ridge_regression", ridge)])
r2, mse = evaluate(estimator, X, y, cv = tscv)
scores.append(np.mean(r2))
plt.plot(alphas, scores)
## regress_test(df, Ridge())
Learning Model
What is the problem here?
from tqdm import tqdm
def Xy(df, target, cls):
if cls:
return df.drop([target, 'Date'], axis=1), df[target] >0
else:
return df.drop([target, 'Date'], axis=1), df[target]
def execute_CV(model, param_grid, X, y, cv, poly = None, gridsearch = True, **kwargs):
if poly != None:
# when both polynomial features and parameter grid are used
scores = {}
poly_able = (X.dtypes != 'uint8').values
X_poly, X_non = X.iloc[:, poly_able], X.iloc[:, ~poly_able]
for i in tqdm(poly):
X2 = PolynomialFeatures(degree=i).fit_transform(X_poly)
X2 = np.concatenate([X2, X_non], axis=1)
if gridsearch:
CV_ = GridSearchCV(model, param_grid, cv=cv, verbose =1, **kwargs)
else:
CV_ = RandomizedSearchCV(model, param_grid, cv=cv, verbose =1, **kwargs)
CV_.fit(X2, y)
scores[CV_.best_score_] = (i, CV_)
mxx = scores[max(scores.keys())]
print(mxx[0])
return mxx[1]
else:
## When only parameter grid are used
if gridsearch:
CV_ = GridSearchCV(model, param_grid, cv=cv, verbose = 1, **kwargs)
else:
CV_ = RandomizedSearchCV(model, param_grid, cv=cv, verbose =1, **kwargs)
CV_.fit(X,y)
print('Best score:', CV_.best_score_)
return CV_
def class_report(y_true, y_pred):
print('Accuracy:', accuracy_score(y_true, y_pred))
print('F1:', f1_score(y_true, y_pred))
return classification_report(y_true, y_pred, output_dict=True)
def measure_error(y_train, y_test, pred_train, pred_test, label):
train = pd.Series({'accuracy':accuracy_score(y_train, pred_train),
'precision': precision_score(y_train, pred_train),
'recall': recall_score(y_train, pred_train),
'f1': f1_score(y_train, pred_train)},
name='train')
test = pd.Series({'accuracy':accuracy_score(y_test, pred_test),
'precision': precision_score(y_test, pred_test),
'recall': recall_score(y_test, pred_test),
'f1': f1_score(y_test, pred_test)},
name='test')
return pd.concat([train, test], axis=1)
from colorsetup import colors, palette
import seaborn as sns
sns.set_palette(palette)
def confusion_plot(y_true, y_pred):
sns.set_palette(sns.color_palette(colors))
_, ax = plt.subplots(figsize=None)
ax = sns.heatmap(confusion_matrix(y_true, y_pred),
annot=True, fmt='d', cmap=colors,
annot_kws={"size": 40, "weight": "bold"})
labels = ['False', 'True']
ax.set_xticklabels(labels, fontsize=25);
ax.set_yticklabels(labels, fontsize=25);
ax.set_ylabel('True value', fontsize=30);
ax.set_xlabel('Prediction', fontsize=30)
return ax
def learning(data: pd.DataFrame, regressor, params = None, clss = False, pred = False,
n_jobs = None, poly = None, scores = None, Date = 'Date', gridsearch = True,
target ='daily_ret', window = 400, pred_window = 15, prnt = True, refit = True):
## training with 6month(120days) and predict 3month(60days)
if pred == True:
data, data_pred = train_test_split(data, test_size=0.1, shuffle = False)
X, y = Xy(data, target, clss)
tscv = TimeSeriesSplit() #n_splits=int(data.shape[0]), max_train_size=window
if params != None:
cvres = execute_CV(regressor, params, X, y, tscv, poly = poly, gridsearch = gridsearch,
scoring= scores, n_jobs = n_jobs, refit = refit)
if pred:
X_pred, y_pred = Xy(data_pred, target, clss)
if prnt:
if clss != True:
print(r2_score(y_pred, cvres.predict(X_pred)))
print(confusion_plot(y_pred>0, cvres.predict(X_pred)>0))
rpt = class_report(y_pred, cvres.predict(X_pred))
return cvres, rpt
else:
return cvres, None
else:
## cross validation only with polynomial features
if poly != None:
scores = {}
poly_able = (X.dtypes != 'uint8').values
X_poly, X_non = X.iloc[:, poly_able], X.iloc[:, ~poly_able]
for i in tqdm(poly):
X2 = PolynomialFeatures(degree=i).fit_transform(X_poly)
X2 = np.concatenate([X2, X_non], axis=1)
cv_results = cross_validate(regressor, X2, y, cv = tscv,
verbose=1)
scores[i] = cv_results
if prnt:
print(scores)
return regressor.fit(X2, y), scores
else:
## no cross validation
res = []
reg = regressor.fit(X, y)
if pred:
X_pred, y_pred = Xy(data_pred, target, clss)
if prnt:
if clss != True:
res.append(r2_score(y_pred, reg.predict(X_pred)))
print(confusion_plot(y_pred>0, reg.predict(X_pred)>0))
else:
res.append(class_report(y_pred, reg.predict(X_pred)))
print(confusion_plot(y_pred>0, reg.predict(X_pred)>0))
else:
res = evaluate(reg, X, y, tscv, clss)
return reg, res
3. Classification
from sklearn.metrics import confusion_matrix, accuracy_score, classification_report, f1_score
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import GradientBoostingClassifier, RandomForestClassifier
3-1 Basic classification models
3-1-1 Logistic Regression
lr = LogisticRegression(solver='liblinear', max_iter= 3000, penalty = 'l1')
params = {
'log__C': np.linspace(10, 40, 5),
}
regressor = Pipeline([
("log", lr)])
reg, rpt = learning(data = df, regressor = regressor, params = params, \
clss = True, pred = True)
res = reg.cv_results_
print(reg.best_params_)
plt.plot(res['param_log__C'], res['mean_test_score'])
Fitting 5 folds for each of 5 candidates, totalling 25 fits
Best score: 0.5893805309734513
Accuracy: 0.6387665198237885
F1: 0.6796875
AxesSubplot(0.125,0.125;0.62x0.755)
{'log__C': 32.5}
[<matplotlib.lines.Line2D at 0x2866f62e0>]
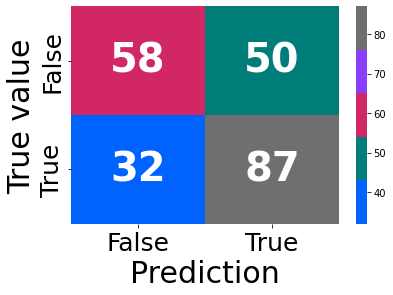
reg
precision recall f1-score support
False 0.56 0.81 0.66 108
True 0.71 0.41 0.52 119
accuracy 0.60 227
macro avg 0.63 0.61 0.59 227
weighted avg 0.64 0.60 0.59 227
3-1-2 Tree based modelm
from io import StringIO
from IPython.display import Image
from sklearn.tree import export_graphviz
import pydotplus
## Estimate dtc model and report outcomes
dt1 = DecisionTreeClassifier()
dt1, rpt = learning(data = df, regressor = dt1,
clss = True, pred = True, prnt = False)
## Estimate dtc model and report outcomes
dtc = DecisionTreeClassifier()
params = {'max_depth':range(1, dt1.tree_.max_depth+1, 2),
'max_features': range(1, len(dt1.feature_importances_)+1)}
reg, rpt = learning(data = df, regressor = dtc, params = params,\
clss = True, pred = True)
print(reg.best_params_)
Fitting 5 folds for each of 639 candidates, totalling 3195 fits
Best score: 0.6466076696165193
AxesSubplot(0.125,0.125;0.62x0.755)
Accuracy: 0.6167400881057269
F1: 0.5538461538461539
{'max_depth': 3, 'max_features': 25}
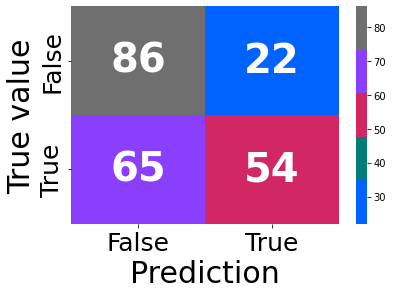
reg.best_par
res = reg.cv_results_
plt.plot(res['param_max_depth'].data, res['mean_test_score'])
[<matplotlib.lines.Line2D at 0x176a475b0>]
#### BEGIN SOLUTION
## Create an output destination for the file
dot_data = StringIO()
export_graphviz(reg.best_estimator_, out_file=dot_data, filled=True)
graph = pydotplus.graph_from_dot_data(dot_data.getvalue())
## View the tree image
filename = 'wine_tree_prune.png'
graph.write_png(filename)
Image(filename=filename)
#### END SOLUTION
3-1-3 KNN
## Estimate KNN model and report outcomes
knn = KNeighborsClassifier()
params = {
'knn__n_neighbors': np.arange(50, 150, 5),
'knn__weights': ['uniform', 'distance'],
}
model = Pipeline([
("knn", knn)])
reg = learning(data = df, regressor = model, params = params,\
clss = True, pred = True)
res = reg.cv_results_
print(reg.best_params_)
Fitting 5 folds for each of 40 candidates, totalling 200 fits
Best score: 0.5386430678466076
precision recall f1-score support
False 0.42 0.24 0.30 106
True 0.51 0.71 0.60 121
accuracy 0.49 227
macro avg 0.47 0.47 0.45 227
weighted avg 0.47 0.49 0.46 227
Accuracy: 0.4889867841409692
F1: 0.5972222222222222
AxesSubplot(0.125,0.125;0.62x0.755)
{'knn__n_neighbors': 145, 'knn__weights': 'uniform'}
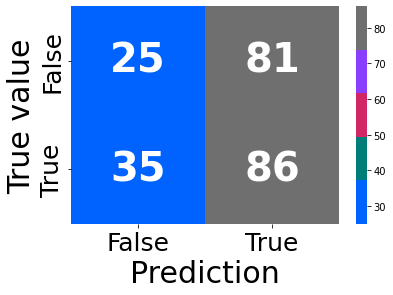
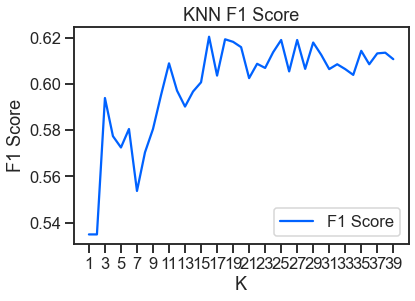
train, test = train_test_split(df, test_size=0.2, shuffle = False)
X_train = train.drop(['daily_ret', 'Date'], axis=1)
y_train = train['daily_ret']>0
X_test = test.drop(['daily_ret','Date'], axis=1)
y_test = test['daily_ret']>0
max_k = 40
f1_scores = list()
error_rates = list() ## 1-accuracy
for k in range(1, max_k):
knn = KNeighborsClassifier(n_neighbors=k, weights='distance')
knn = knn.fit(X_train, y_train)
y_pred = knn.predict(X_test)
f1 = f1_score(y_pred, y_test)
f1_scores.append((k, round(f1_score(y_test, y_pred), 4)))
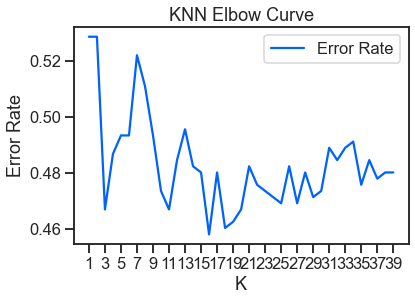
error = 1-round(accuracy_score(y_test, y_pred), 4)
error_rates.append((k, error))
f1_results = pd.DataFrame(f1_scores, columns=['K', 'F1 Score'])
error_results = pd.DataFrame(error_rates, columns=['K', 'Error Rate'])
## Plot F1 results
sns.set_context('talk')
sns.set_style('ticks')
plt.figure(dpi=300)
ax = f1_results.set_index('K').plot(color=colors[0])
ax.set(xlabel='K', ylabel='F1 Score')
ax.set_xticks(range(1, max_k, 2));
plt.title('KNN F1 Score')
## plt.savefig('knn_f1.png')
Text(0.5, 1.0, 'KNN F1 Score')
<Figure size 1800x1200 with 0 Axes>
## Plot Accuracy (Error Rate) results
sns.set_context('talk')
sns.set_style('ticks')
plt.figure(dpi=300)
ax = error_results.set_index('K').plot(color=colors[0])
ax.set(xlabel='K', ylabel='Error Rate')
ax.set_xticks(range(1, max_k, 2))
plt.title('KNN Elbow Curve')
Text(0.5, 1.0, 'KNN Elbow Curve')
<Figure size 1800x1200 with 0 Axes>
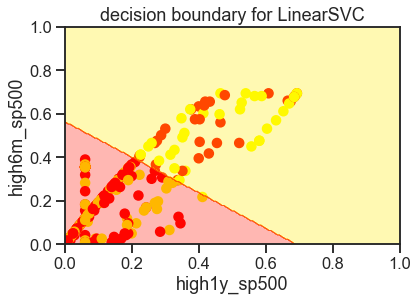
3-2 Linear Decision boundary
from sklearn.svm import LinearSVC
X = df.drop(['daily_ret', 'Date'], axis=1)
y = df['daily_ret']>0
fields = list(X.columns[:-1])
correlations = abs(X[fields].corrwith(y))
correlations.sort_values(inplace=True)
fields = correlations.map(abs).sort_values().iloc[-2:].index
X_sub = X[fields]
LSVC = LinearSVC()
LSVC.fit(X_sub.values, y)
X_color = X_sub.sample(300, random_state=45)
y_color = y.loc[X_color.index]
y_color = y_color.map(lambda r: 'red' if r == 1 else 'yellow')
ax = plt.axes()
ax.scatter(
X_color.iloc[:, 0], X_color.iloc[:, 1],
color=y_color, alpha=1)
## -----------
x_axis, y_axis = np.arange(0, 1.005, .005), np.arange(0, 1.005, .005)
xx, yy = np.meshgrid(x_axis, y_axis)
xx_ravel = xx.ravel()
yy_ravel = yy.ravel()
X_grid = pd.DataFrame([xx_ravel, yy_ravel]).T
y_grid_predictions = LSVC.predict(X_grid)
y_grid_predictions = y_grid_predictions.reshape(xx.shape)
ax.contourf(xx, yy, y_grid_predictions, cmap=plt.cm.autumn_r, alpha=.3)
## -----------
ax.set(
xlabel=fields[0],
ylabel=fields[1],
xlim=[0, 1],
ylim=[0, 1],
title='decision boundary for LinearSVC');
def plot_decision_boundary(estimator, X, y):
estimator.fit(X.values, y)
X_color = X.sample(300)
y_color = y.loc[X_color.index]
y_color = y_color.map(lambda r: 'red' if r == 1 else 'blue')
x_axis, y_axis = np.arange(0, 1, .005), np.arange(0, 1, .005)
xx, yy = np.meshgrid(x_axis, y_axis)
xx_ravel = xx.ravel()
yy_ravel = yy.ravel()
X_grid = pd.DataFrame([xx_ravel, yy_ravel]).T
y_grid_predictions = estimator.predict(X_grid.values)
y_grid_predictions = y_grid_predictions.reshape(xx.shape)
fig, ax = plt.subplots(figsize=(10, 10))
ax.contourf(xx, yy, y_grid_predictions, cmap=plt.cm.autumn_r, alpha=.1)
ax.scatter(X_color.iloc[:, 0], X_color.iloc[:, 1], color=y_color, alpha=1)
ax.set(
xlabel=fields[0],
ylabel=fields[1],
title=str(estimator))
from sklearn.svm import SVC
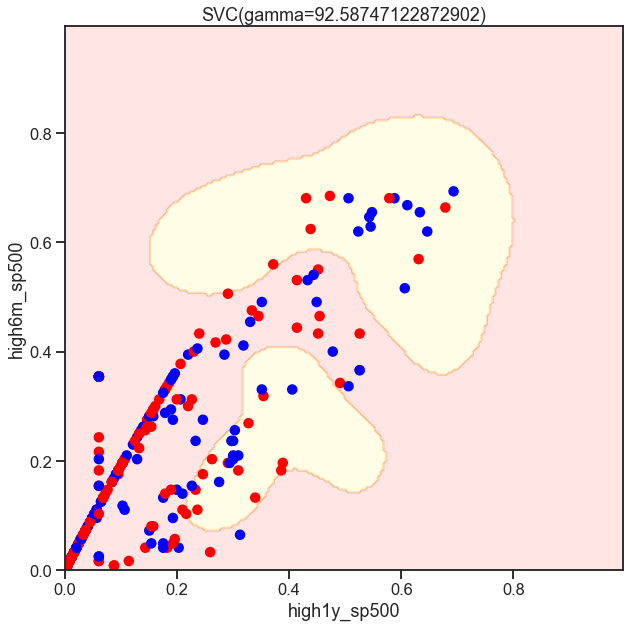
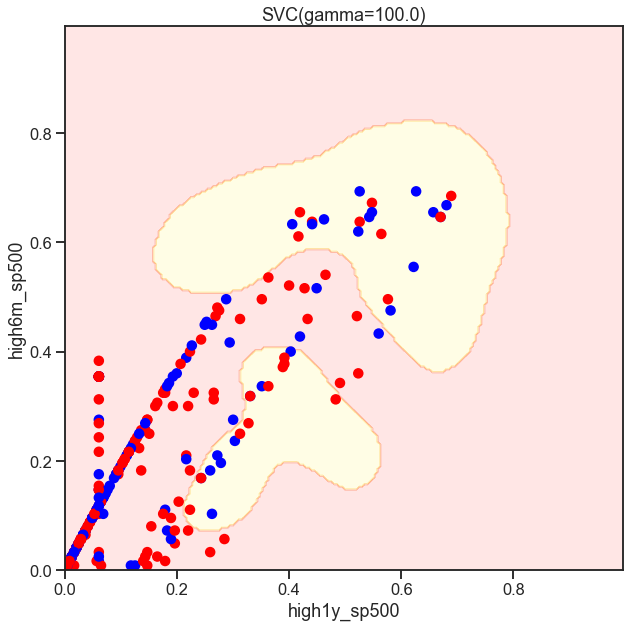
gammas = np.geomspace(50, 100, num=10)
for gamma in gammas:
SVC_Gaussian = SVC(kernel='rbf', gamma=gamma)
plot_decision_boundary(SVC_Gaussian, X_sub, y)
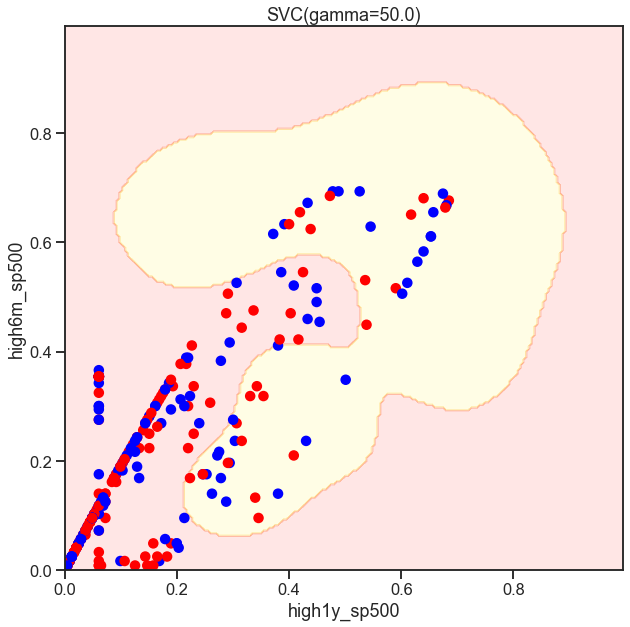
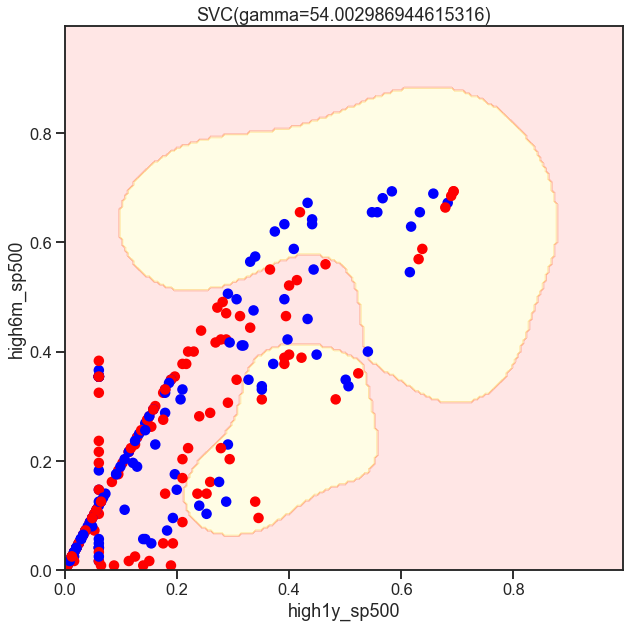
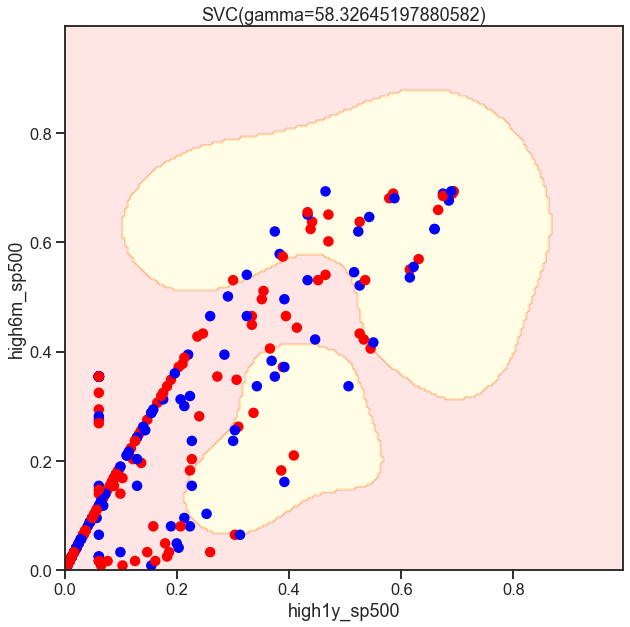
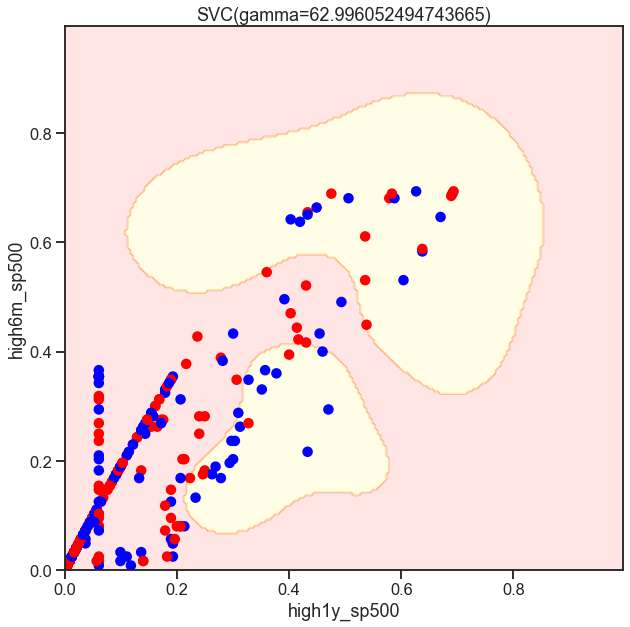
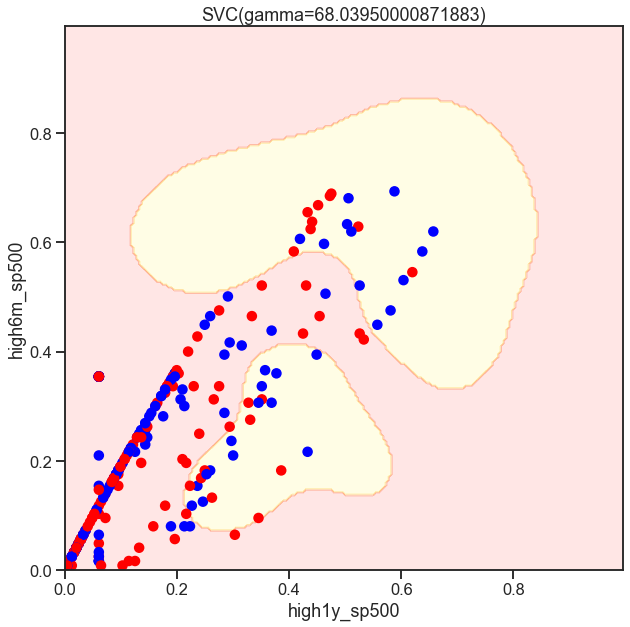
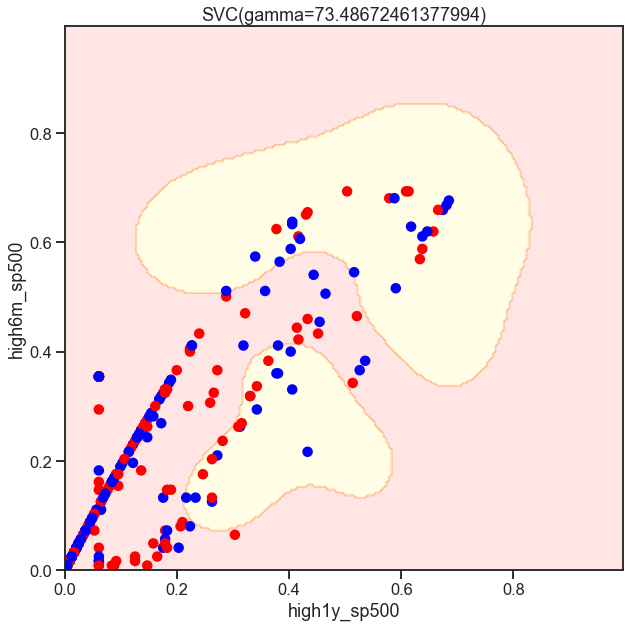
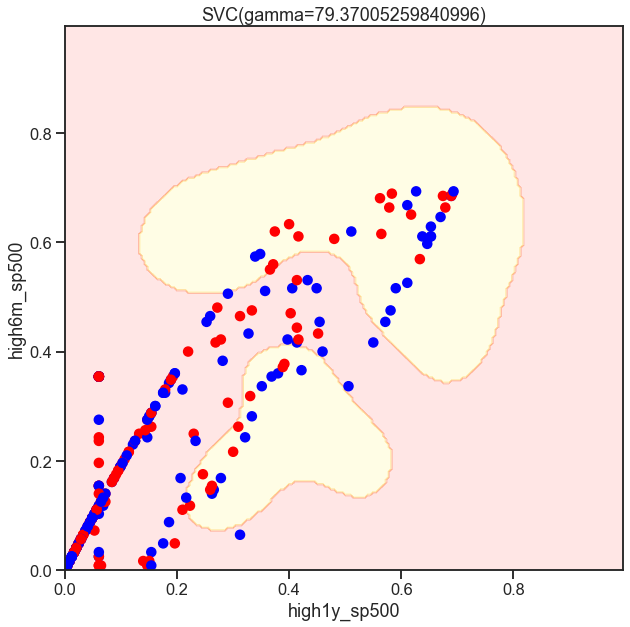
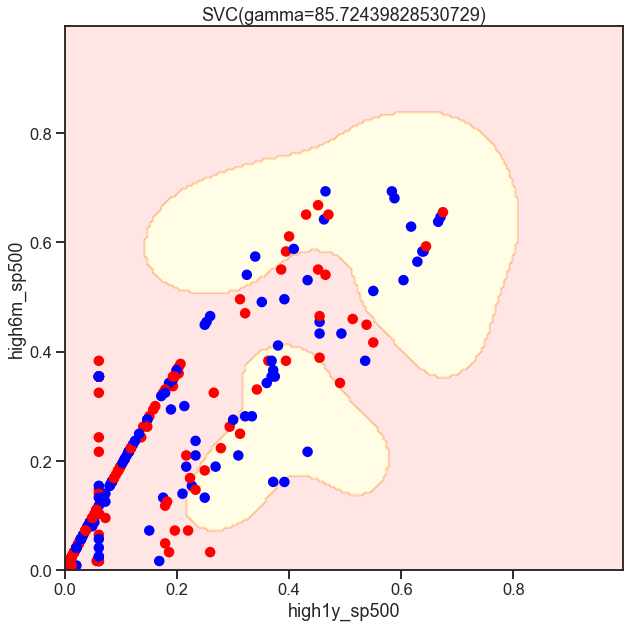
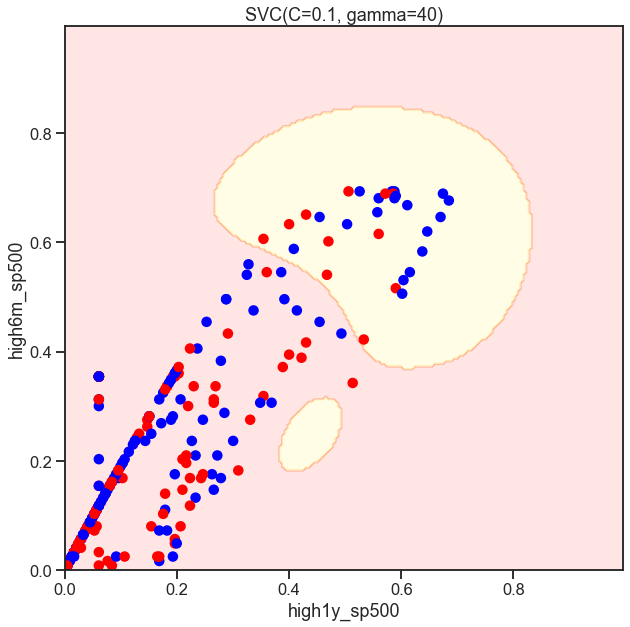
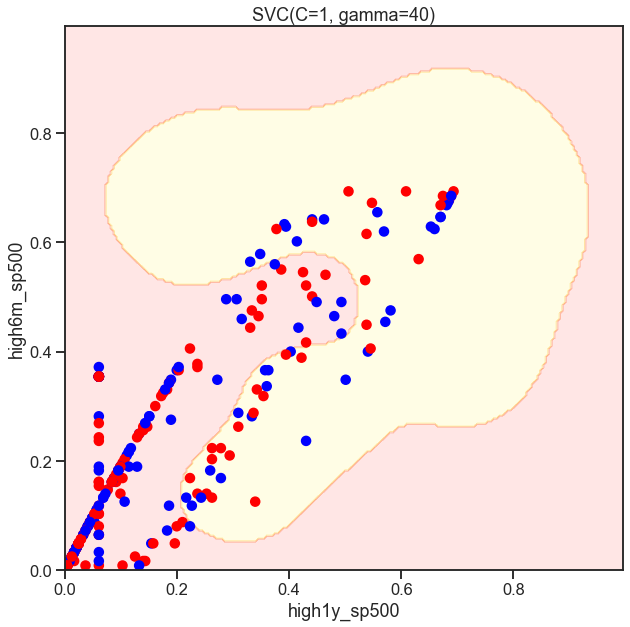
Cs = [.1, 1, 10]
for C in Cs:
SVC_Gaussian = SVC(kernel='rbf', gamma=40, C=C)
plot_decision_boundary(SVC_Gaussian, X, y)
/Users/hun/miniforge3/envs/hun/lib/python3.9/site-packages/sklearn/base.py:450: UserWarning: X does not have valid feature names, but SVC was fitted with feature names
warnings.warn(
/Users/hun/miniforge3/envs/hun/lib/python3.9/site-packages/sklearn/base.py:450: UserWarning: X does not have valid feature names, but SVC was fitted with feature names
warnings.warn(
/Users/hun/miniforge3/envs/hun/lib/python3.9/site-packages/sklearn/base.py:450: UserWarning: X does not have valid feature names, but SVC was fitted with feature names
warnings.warn(
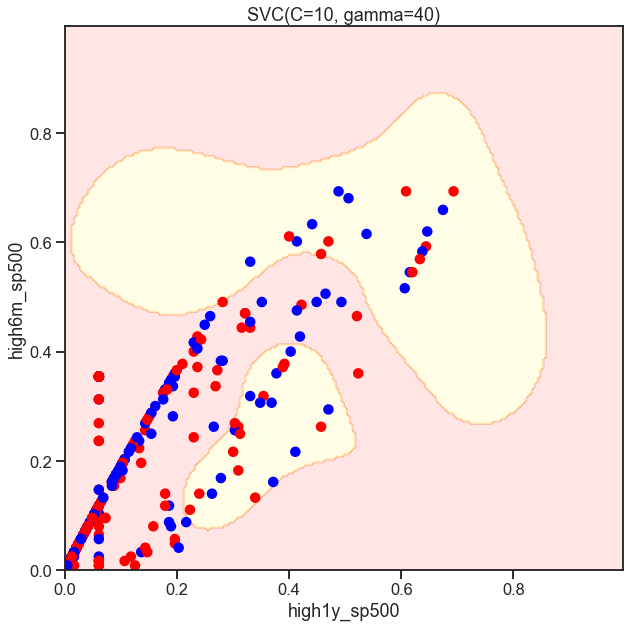
Classifcation
- SVC, nystroem converge time
- Tree model
from sklearn.kernel_approximation import Nystroem
from sklearn.svm import SVC
from sklearn.linear_model import SGDClassifier
X = df.drop(['daily_ret', 'Date'], axis=1)
kwargs = {'kernel': 'rbf'}
svc = SVC(**kwargs)
nystroem = Nystroem(**kwargs)
sgd = SGDClassifier()
%%timeit
svc.fit(X, y)
158 ms ± 894 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)
%%timeit
X_transformed = nystroem.fit_transform(X)
sgd.fit(X_transformed, y)
50.2 ms ± 15.8 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
%%timeit
X2_transformed = nystroem.fit_transform(X2)
sgd.fit(X2_transformed, y2)
train_test_full_error
#### END SOLUTION
| train | test | |
|---|---|---|
| accuracy | 1.0 | 0.513216 |
| precision | 1.0 | 0.522388 |
| recall | 1.0 | 0.600858 |
| f1 | 1.0 | 0.558882 |
from sklearn.model_selection import GridSearchCV
param_grid = {'max_depth':range(1, dt.tree_.max_depth+1, 2),
'max_features': range(1, len(dt.feature_importances_)+1)}
GR = GridSearchCV(DecisionTreeClassifier(random_state=42),
param_grid=param_grid,
scoring='recall',
n_jobs=-1)
GR = GR.fit(X_train, y_train)
GR.best_estimator_.tree_.node_count, GR.best_estimator_.tree_.max_depth
(3, 1)
y_train_pred_gr = GR.predict(X_train)
y_test_pred_gr = GR.predict(X_test)
train_test_gr_error = pd.concat([measure_error(y_train, y_train_pred_gr, 'train'),
measure_error(y_test, y_test_pred_gr, 'test')],
axis=1)
train_test_gr_error
| train | test | |
|---|---|---|
| accuracy | 0.529801 | 0.513216 |
| precision | 0.529801 | 0.513216 |
| recall | 1.000000 | 1.000000 |
| f1 | 0.692641 | 0.678311 |
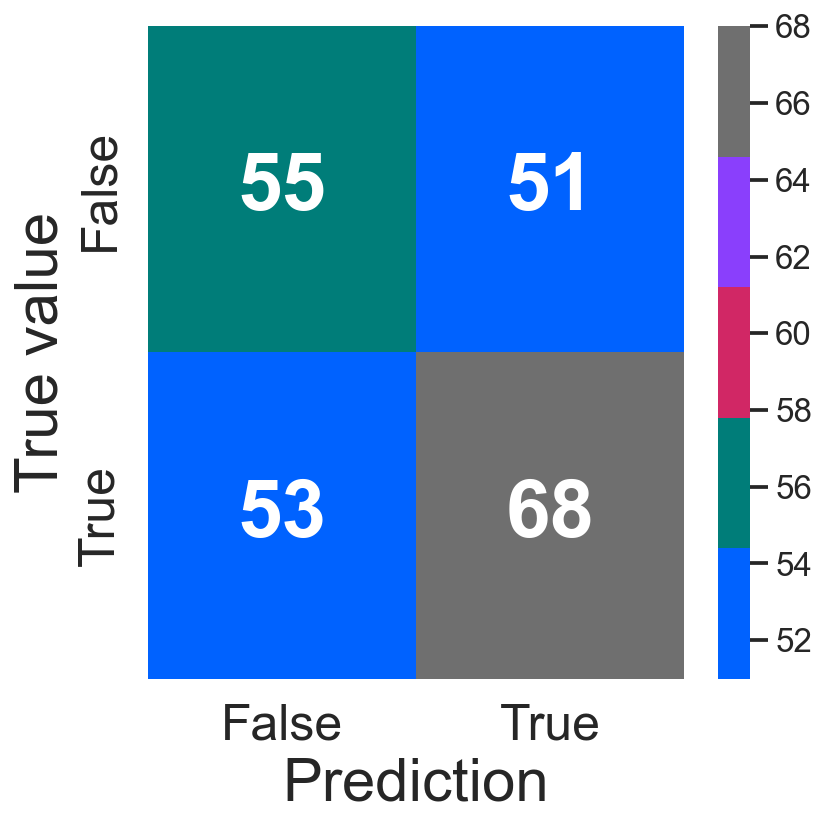
confusion_plot(y_test, y_test_pred_gr)
<AxesSubplot:xlabel='Prediction', ylabel='True value'>
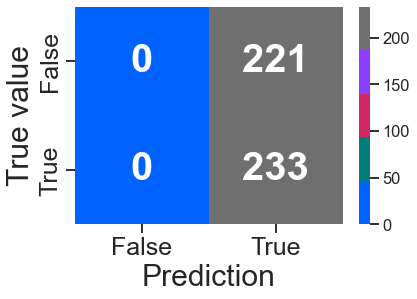
3-3 Classification Ensamble
## Estimate dtc model and report outcomes
gbc = GradientBoostingClassifier(random_state=777, n_iter_no_change= 10)
gbc, rpt = learning(data = df, regressor = gbc, clss = True, pred = True, prnt = False)
gbc2 = GradientBoostingClassifier(random_state=777, n_iter_no_change = 10, n_estimators= gbc.n_estimators_+10)
params = {'max_depth':range(3, len(gbc.feature_importances_), 2),
'max_features': range(1, gbc.max_features_+1)}
reg, rpt = learning(data = df, regressor = gbc2, params = params, clss = True, pred = True)
res = reg.cv_results_
print(reg.best_params_)
Fitting 5 folds for each of 2414 candidates, totalling 12070 fits
Best score: 0.6525073746312684
AxesSubplot(0.125,0.125;0.62x0.755)
Accuracy: 0.5903083700440529
F1: 0.541871921182266
{'max_depth': 3, 'max_features': 45}
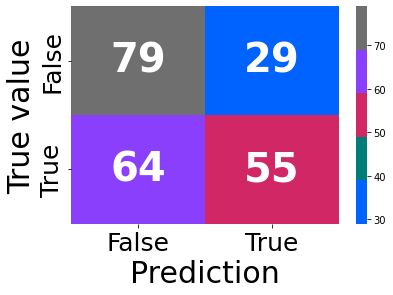
model = reg.best_estimator_
feature_imp = pd.Series(model.feature_importances_, index=df.drop(['Date','daily_ret'],axis=1).columns).sort_values(ascending=False)
ax = feature_imp.plot(kind='bar', figsize=(16, 6))
ax.set(ylabel='Relative Importance');
ax.set(ylabel='Feature');

4. Unsupervised
aapl2 = pd.read_csv('./data/aapl2.csv').dropna()
## float_columns = [i for i in aapl2.columns if i not in ['Date']]
## no_cols = [i for i in aapl.columns if i not in float_columns]
## aapl2 = pd.merge(aapl[no_cols], aapl2, on='Date', how = 'left')
float_columns = [i for i in aapl2.columns if aapl2.dtypes[i] != object]
## sets backend to render higher res images
%config InlineBackend.figure_formats = ['retina']
import numpy as np, pandas as pd, seaborn as sns, matplotlib.pyplot as plt
from sklearn.preprocessing import scale, StandardScaler, MinMaxScaler
from sklearn.cluster import KMeans, AgglomerativeClustering
4-1 K-means clustering
plt.rcParams['figure.figsize'] = [6,6]
sns.set_style("whitegrid")
sns.set_context("talk")
## helper function that allows us to display data in 2 dimensions an highlights the clusters
## def display_cluster(X,kmeans):
## plt.scatter(X[:,0],
## X[:,1])
## ## Plot the clusters
## plt.scatter(kmeans.cluster_centers_[:, 0],
## kmeans.cluster_centers_[:, 1],
## s=200, ## Set centroid size
## c='red') ## Set centroid color
## plt.show()
def display_cluster(X,km=[],num_clusters=0):
color = 'brgcmyk'
alpha = 0.5
s = 20
if num_clusters == 0:
plt.scatter(X[:,0],X[:,1],c = color[0],alpha = alpha,s = s)
else:
for i in range(num_clusters):
plt.scatter(X[km.labels_==i,0],X[km.labels_==i,1],c = color[i],alpha = alpha,s=s)
plt.scatter(km.cluster_centers_[i][0],km.cluster_centers_[i][1],c = color[i], marker = 'x', s = 100)
X_cdf = aapl2[['Close_sp500', 'Close_vix']]
cdf = StandardScaler().fit(X_cdf).transform(X_cdf)
num_clusters = 5
km = KMeans(n_clusters=num_clusters, n_init=10) ## n_init, number of times the K-mean algorithm will run
km.fit(cdf)
display_cluster(cdf, km, num_clusters=num_clusters)
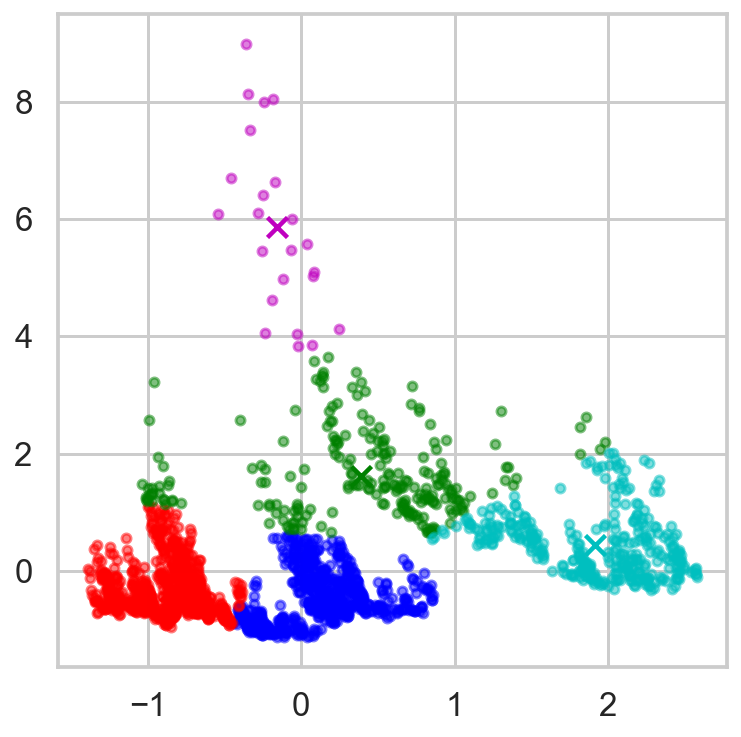
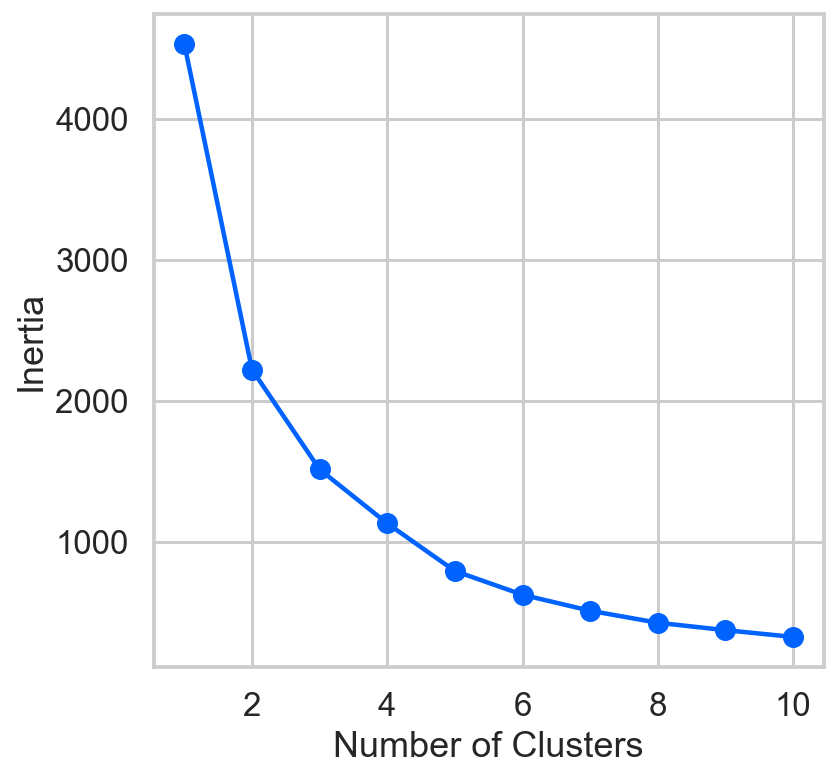
inertia = []
list_num_clusters = list(range(1,11))
for num_clusters in list_num_clusters:
km = KMeans(n_clusters=num_clusters)
km.fit(cdf)
inertia.append(km.inertia_)
plt.plot(list_num_clusters,inertia)
plt.scatter(list_num_clusters,inertia)
plt.xlabel('Number of Clusters')
plt.ylabel('Inertia');
4-2 Comparing in multidimension
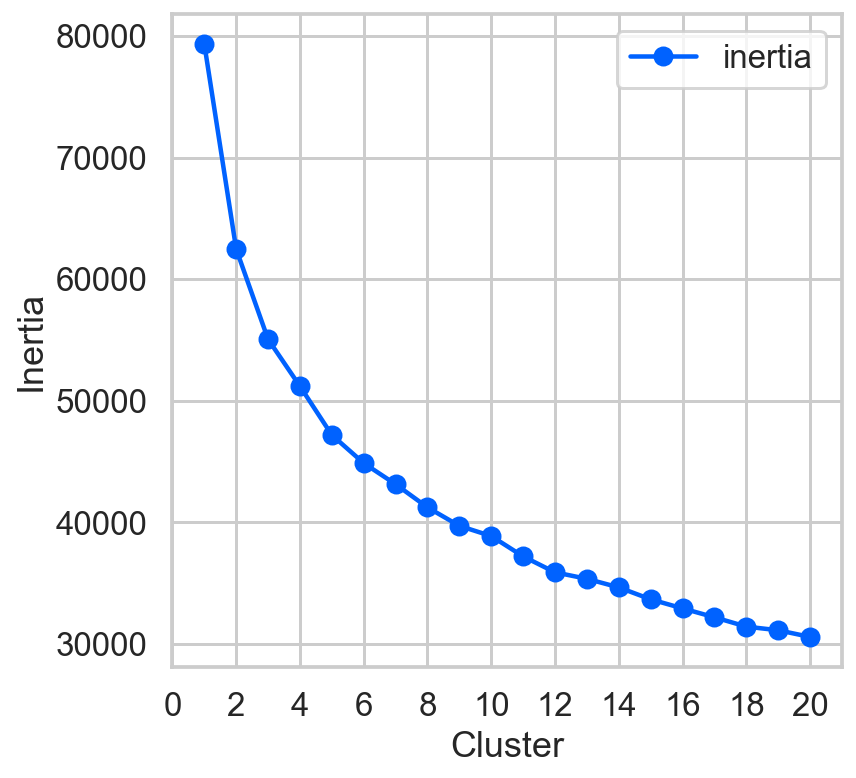
km_list = list()
data = StandardScaler().fit(aapl2[float_columns]).transform(aapl2[float_columns])
for clust in range(1,21):
km = KMeans(n_clusters=clust, random_state=42)
km = km.fit(data)
km_list.append(pd.Series({'clusters': clust,
'inertia': km.inertia_,
'model': km}))
plot_data = (pd.concat(km_list, axis=1).T\
[['clusters','inertia']].set_index('clusters'))
ax = plot_data.plot(marker='o',ls='-')
ax.set_xticks(range(0,21,2))
ax.set_xlim(0,21)
ax.set(xlabel='Cluster', ylabel='Inertia');
/Users/hun/miniforge3/envs/hun/lib/python3.9/site-packages/pandas/core/indexes/base.py:6982: FutureWarning: In a future version, the Index constructor will not infer numeric dtypes when passed object-dtype sequences (matching Series behavior)
return Index(sequences[0], name=names)
res = pd.DataFrame()
res['daily_ret'] = aapl2.daily_ret
data = StandardScaler().fit(aapl2[float_columns]).transform(aapl2[float_columns])
km = KMeans(n_clusters=5, n_init=20, random_state=777)
km = km.fit(data)
res['kmeans'] = km.predict(data)
ag = AgglomerativeClustering(n_clusters=5, linkage='ward', compute_full_tree=True)
ag = ag.fit(data)
res['agglom'] = ag.fit_predict(data)
res2 = (res[['daily_ret','agglom','kmeans']]
.groupby(['daily_ret','agglom','kmeans'])
.size()
.to_frame()
.rename(columns={0:'number'}))
print(res2.to_latex())
\begin{tabular}{lllr}
\toprule
& & & number \\
daily\_ret & agglom & kmeans & \\
\midrule
Bear & 0 & 0 & 215 \\
& & 1 & 43 \\
& & 2 & 1 \\
& & 3 & 22 \\
& & 4 & 1 \\
& 1 & 0 & 6 \\
& & 2 & 2 \\
& & 4 & 18 \\
& 2 & 0 & 3 \\
& & 1 & 88 \\
& & 2 & 1 \\
& & 3 & 266 \\
& 3 & 2 & 230 \\
& & 4 & 4 \\
& 4 & 0 & 6 \\
& & 1 & 163 \\
Bull & 0 & 0 & 203 \\
& & 1 & 40 \\
& & 3 & 21 \\
& & 4 & 1 \\
& 1 & 0 & 4 \\
& & 2 & 1 \\
& & 4 & 13 \\
& 2 & 0 & 13 \\
& & 1 & 114 \\
& & 3 & 339 \\
& 3 & 0 & 2 \\
& & 2 & 266 \\
& & 4 & 3 \\
& 4 & 0 & 8 \\
& & 1 & 170 \\
\bottomrule
\end{tabular}
/var/folders/1t/_7p_zm4x449blqs7bvqvb0rm0000gn/T/ipykernel_66451/732263353.py:19: FutureWarning: In future versions `DataFrame.to_latex` is expected to utilise the base implementation of `Styler.to_latex` for formatting and rendering. The arguments signature may therefore change. It is recommended instead to use `DataFrame.style.to_latex` which also contains additional functionality.
print(res2.to_latex())
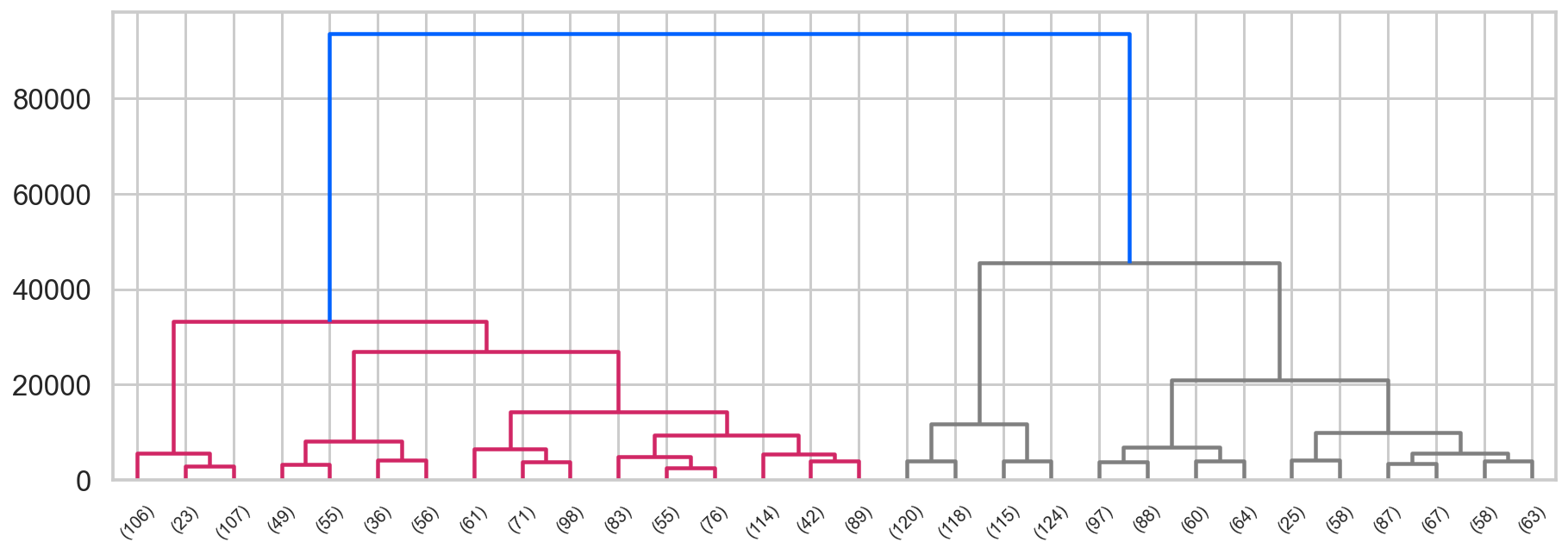
## First, we import the cluster hierarchy module from SciPy (described above) to obtain the linkage and dendrogram functions.
from scipy.cluster import hierarchy
## Some color setup
red,blue = colors[2], colors[0]
Z = hierarchy.linkage(ag.children_, method='ward')
fig, ax = plt.subplots(figsize=(15,5))
hierarchy.set_link_color_palette([red, 'gray'])
den = hierarchy.dendrogram(Z, orientation='top',
p=30, truncate_mode='lastp',
show_leaf_counts=True, ax=ax,
above_threshold_color=blue)
4-3 utilizing in regression
X_cdf = aapl2[['Close_sp500', 'Close_vix', 'Close_bond_2y']]
data = StandardScaler().fit(X_cdf).transform(X_cdf)
km = KMeans(n_clusters=5, n_init=20, random_state=777)
km = km.fit(data)
aapl['kmeans'] = km.predict(data)[1:]
#### BEGIN SOLUTION
ag = AgglomerativeClustering(n_clusters=5, linkage='ward', compute_full_tree=True)
ag = ag.fit(data)
aapl['agglom'] = ag.fit_predict(data)[1:]
one_hot_encode_cols = ['kmeans','agglom'] ## list of categorical fields
df = pd.get_dummies(aapl, columns=one_hot_encode_cols, drop_first=True).reset_index(drop=True)
## Estimate dtc model and report outcomes
gbc = GradientBoostingClassifier(random_state=777, n_iter_no_change= 10)
gbc = learning(data = df, regressor = gbc, clss = True, pred = True, prnt = False)
gbc2 = GradientBoostingClassifier(random_state=777, n_iter_no_change = 10, n_estimators= gbc.n_estimators_+10)
params = {'max_depth':range(3, len(gbc.feature_importances_), 2),
'max_features': range(1, gbc.max_features_+1)}
reg = learning(data = df, regressor = gbc2, params = params, clss = True, pred = True)
res = reg.cv_results_
print(reg.best_params_)
Fitting 5 folds for each of 1890 candidates, totalling 9450 fits
Best score: 0.5398230088495575
precision recall f1-score support
False 0.51 0.52 0.51 106
True 0.57 0.56 0.57 121
accuracy 0.54 227
macro avg 0.54 0.54 0.54 227
weighted avg 0.54 0.54 0.54 227
Accuracy: 0.5418502202643172
F1: 0.5666666666666667
AxesSubplot(0.125,0.125;0.62x0.755)
{'max_depth': 17, 'max_features': 17}
4-4 PCA
from sklearn.decomposition import PCA
pca_list = list()
feature_weight_list = list()
## Fit a range of PCA models
data = StandardScaler().fit(aapl2[float_columns]).transform(aapl2[float_columns])
for n in range(6, 10):
## Create and fit the model
PCAmod = PCA(n_components=n)
PCAmod.fit(data)
## Store the model and variance
pca_list.append(pd.Series({'n':n, 'model':PCAmod,
'var': PCAmod.explained_variance_ratio_.sum()}))
## Calculate and store feature importances
abs_feature_values = np.abs(PCAmod.components_).sum(axis=0)
feature_weight_list.append(pd.DataFrame({'n':n,
'features': float_columns,
'values':abs_feature_values/abs_feature_values.sum()}))
pca_df = pd.concat(pca_list, axis=1).T.set_index('n')
features_df = (pd.concat(feature_weight_list)
.pivot(index='n', columns='features', values='values'))
features_df
/Users/hun/miniforge3/envs/hun/lib/python3.9/site-packages/pandas/core/indexes/base.py:6982: FutureWarning: In a future version, the Index constructor will not infer numeric dtypes when passed object-dtype sequences (matching Series behavior)
return Index(sequences[0], name=names)
| features | AAPL_factor1 | AAPL_factor2 | CO | Close | Close_bond_10y | Close_bond_1m | Close_bond_1y | Close_bond_2y | Close_itw | Close_nasdaq | ... | ma120 | ma20 | std120 | std20 | std_high20 | std_intra | std_open20 | support_high | support_low | support_open |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | |||||||||||||||||||||
| 6 | 0.030236 | 0.037193 | 0.008199 | 0.027970 | 0.032612 | 0.032992 | 0.033682 | 0.034586 | 0.026851 | 0.027899 | ... | 0.032207 | 0.030129 | 0.018837 | 0.018717 | 0.018823 | 0.016651 | 0.019013 | 0.034963 | 0.035248 | 0.037478 |
| 7 | 0.032490 | 0.032149 | 0.010461 | 0.025679 | 0.029585 | 0.028476 | 0.028742 | 0.029950 | 0.026914 | 0.027032 | ... | 0.028294 | 0.029327 | 0.019782 | 0.021357 | 0.021676 | 0.026183 | 0.021668 | 0.032671 | 0.031947 | 0.034375 |
| 8 | 0.036669 | 0.028255 | 0.012337 | 0.022213 | 0.029923 | 0.028524 | 0.029117 | 0.030328 | 0.025168 | 0.023790 | ... | 0.031052 | 0.026635 | 0.018100 | 0.022617 | 0.022573 | 0.024953 | 0.022605 | 0.030334 | 0.029549 | 0.031957 |
| 9 | 0.037665 | 0.028583 | 0.028795 | 0.021349 | 0.029740 | 0.027302 | 0.028086 | 0.029355 | 0.023119 | 0.022945 | ... | 0.033827 | 0.024899 | 0.017160 | 0.021789 | 0.021669 | 0.024982 | 0.021553 | 0.029501 | 0.029063 | 0.031652 |
4 rows × 35 columns
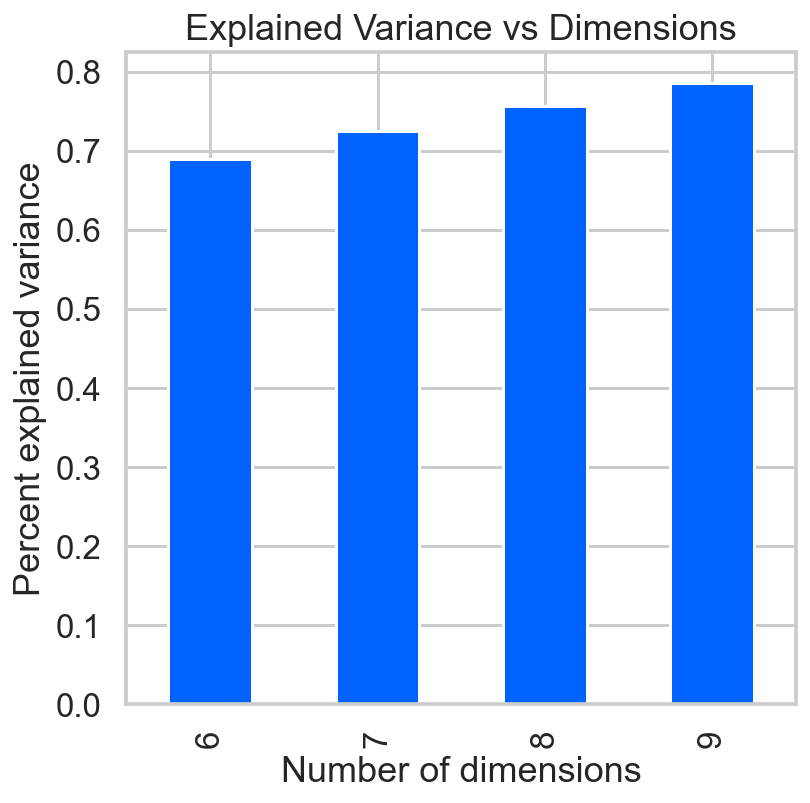
sns.set_context('talk')
ax = pca_df['var'].plot(kind='bar')
ax.set(xlabel='Number of dimensions',
ylabel='Percent explained variance',
title='Explained Variance vs Dimensions');
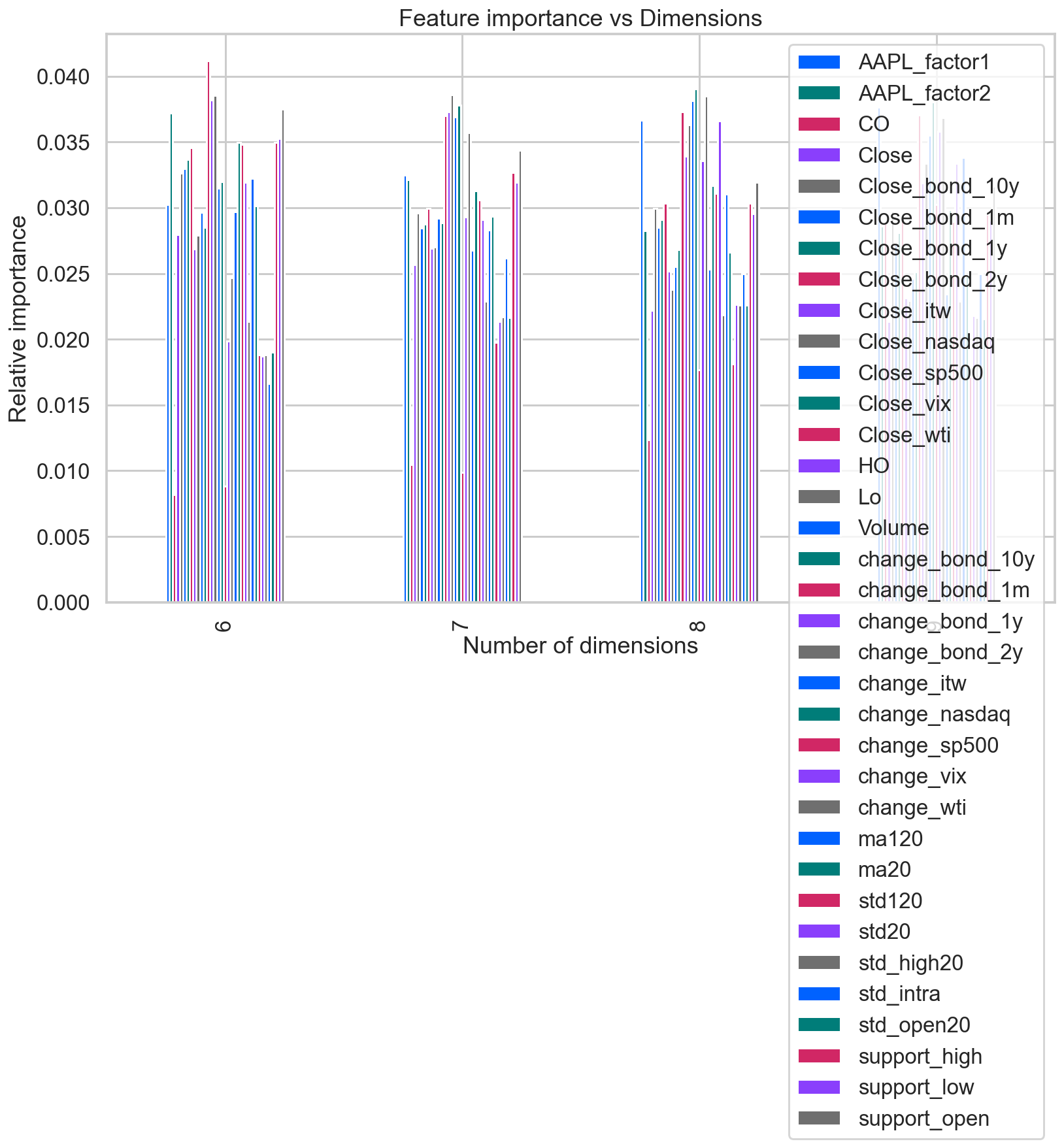
ax = features_df.plot(kind='bar', figsize=(13,8))
ax.legend(loc='upper right')
ax.set(xlabel='Number of dimensions',
ylabel='Relative importance',
title='Feature importance vs Dimensions');
from sklearn.decomposition import KernelPCA
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import mean_squared_error
## Custom scorer--use negative rmse of inverse transform
def scorer(pcamodel, X, y=None):
try:
X_val = X.values
except:
X_val = X
## Calculate and inverse transform the data
data_inv = pcamodel.fit(X_val).transform(X_val)
data_inv = pcamodel.inverse_transform(data_inv)
## The error calculation
mse = mean_squared_error(data_inv.ravel(), X_val.ravel())
## Larger values are better for scorers, so take negative value
return -1.0 * mse
## The grid search parameters
param_grid = {'gamma':[0.001, 0.01, 0.05, 0.1, 0.5, 1.0],
'n_components': [2, 3, 4]}
## The grid search
kernelPCA = GridSearchCV(KernelPCA(kernel='rbf', fit_inverse_transform=True),
param_grid=param_grid,
scoring=scorer,
n_jobs=-1)
kernelPCA = kernelPCA.fit(data)
kernelPCA.best_estimator_
KernelPCA(fit_inverse_transform=True, gamma=0.05, kernel='rbf', n_components=4)
kernelPCA.best_estimator_.eigenvalues_
array([163.71226988, 144.0926473 , 93.50586323, 65.88489674])
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score
X = aapl2[float_columns]
y = aapl2.daily_ret
sss = TimeSeriesSplit(n_splits=5)
def get_avg_score(n):
pipe = [
('scaler', StandardScaler()),
('pca', PCA(n_components=n)),
('estimator', LogisticRegression(solver='liblinear'))
]
pipe = Pipeline(pipe)
scores = []
for train_index, test_index in sss.split(X, y):
global X_train, y_train
X_train, X_test = X.iloc[train_index], X.iloc[test_index]
y_train, y_test = y.iloc[train_index], y.iloc[test_index]
pipe.fit(X_train, y_train)
scores.append(accuracy_score(y_test, pipe.predict(X_test)))
return np.mean(scores)
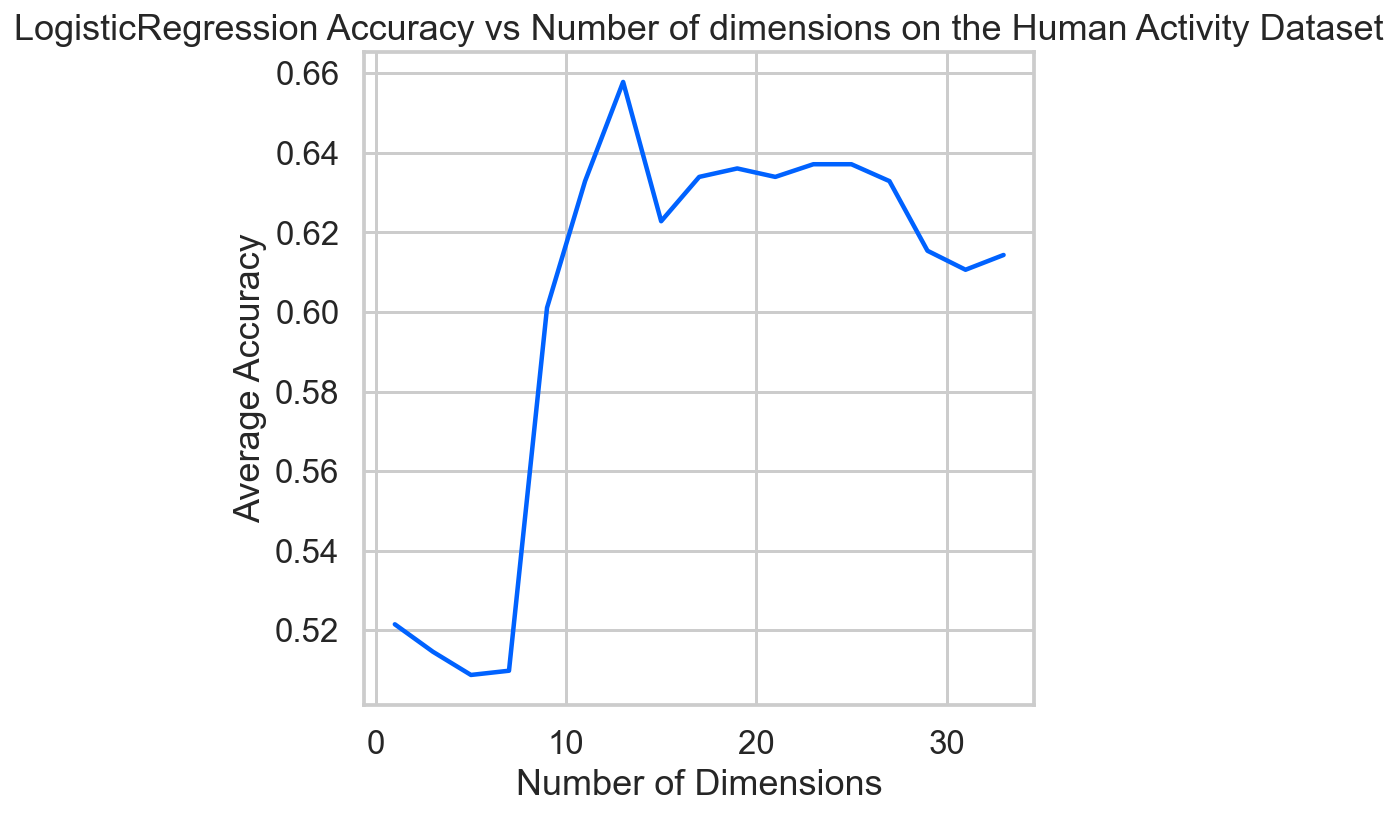
ns = np.arange(1, 35, 2)
score_list = [get_avg_score(n) for n in ns]
sns.set_context('talk')
ax = plt.axes()
ax.plot(ns, score_list)
ax.set(xlabel='Number of Dimensions',
ylabel='Average Accuracy',
title='LogisticRegression Accuracy vs Number of dimensions on the Human Activity Dataset')
ax.grid(True)
ETC
### How to make pipeline by features?
from sklearn.pipeline import make_union, make_pipeline
from sklearn.preprocessing import FunctionTransformer
def get_text_cols(df):
return df[['name', 'fruit']]
def get_num_cols(df):
return df[['height','age']]
vec = make_union(*[
make_pipeline(FunctionTransformer(get_text_cols, validate=False), LabelEncoder()))),
make_pipeline(FunctionTransformer(get_num_cols, validate=False), MinMaxScaler())))
])

Leave a comment