Dimensionality Reduction
Tags: PCA, Unsupervised, Week3
Categories: IBM Machine Learning
Updated:
Dimensionality Reduction
Too many features leads to worse performance. Distance measures perform poorly and the indicent of outliers increases. Data can be represented in a lower dimensional space. Reduce dimensionality by selecting subset (feature elimination). Combine with linear and non-linear transformation.
PCA
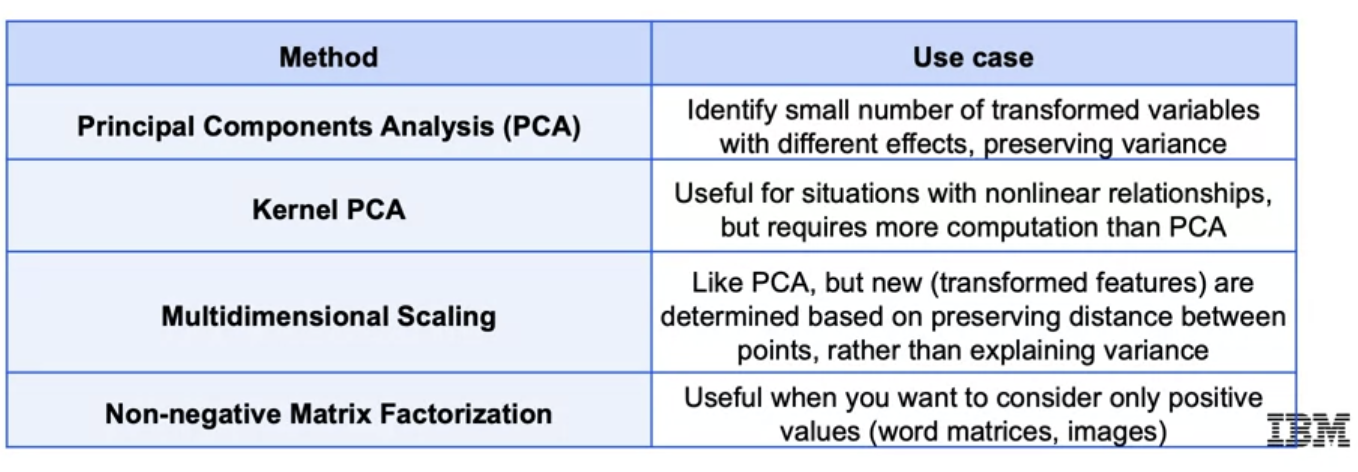
Principal Component Analysis (PCA) is a dimensionality reduction technique. It is a linear transformation that projects the data into a lower dimensional space. Let direction and length be the first principal component. is perpendicular to which has length .
SVD
Principal components are calculated from
Truncated SVD is used for dimensionality
reduction from n to k
Variance is sensitive to scaling .
from sklearn.decompositon import PCA
PCAinst = PCA(n_components=2) #create instance
x_trans = PCAinst.fit_transform(x_train) #fit the instance on the data
Non-linear
Kernel PCA
Use kernel trick introduced in SVM to map down linear relationship.
from sklearn.decomposition import KernelPCA
kPCA = KernelPCA(n_components=2, kernel='rbf', gamma=0.04)
x_kpca = KPCA.fit_transform(x_train)
Multi-Dimensional Scaling (MDS)
MDS maintains the distance between points in a low-dimensional space.
from sklearn.decomposition import MDS
mds = MDS(n_components=2)
x_mds = mds.fit_transform(x_train)
Others: Isomap, TSNE
Non-negative Matrix Factorization
Decompositing matrix only non-negative values. For example,
vectorized words, images. Let
so that
. Also in Image, we can compress only shaded values.
Can never undo the application of a latent feature, it is much
more careful about what it adds at each step. In some
applications, this can make for more human interpretable latent
features.
Thus NMF are non othogonal.
from sklearn.decomposition import NMF
nmf = NMF(n_components=2, init='random')
x_nmf = nmf.fit(x)
Summary

Leave a comment